dRenSeq
Late Blight

In the EU, almost 6m hectares of potatoes with a value of €6 million are grown. Globally, Potato Blight is responsible for €6 billion losses in terms of costs of control and damage.
The most effective chemicals farmers rely on to control blight are being withdrawn and creating new potato varieties, with durable resistance to late blight, is critical for protecting the future of the industry.
Breeders need a tool to accurately determine which known resistance genes are present in breeding material to identify new resistances and to generate novel resistance gene combinations.
The Pathogen
Late blight is constantly evolving and spreading wherever potatoes and tomatoes grow. It’s no surprise that it's name Phytophthora infestans means plant destroyer in Greek.
Each late blight isolate possesses a cocktail of proteins used to fight the plant’s defences and the challenge facing scientists is to discover the proteins that are conserved across world populations.
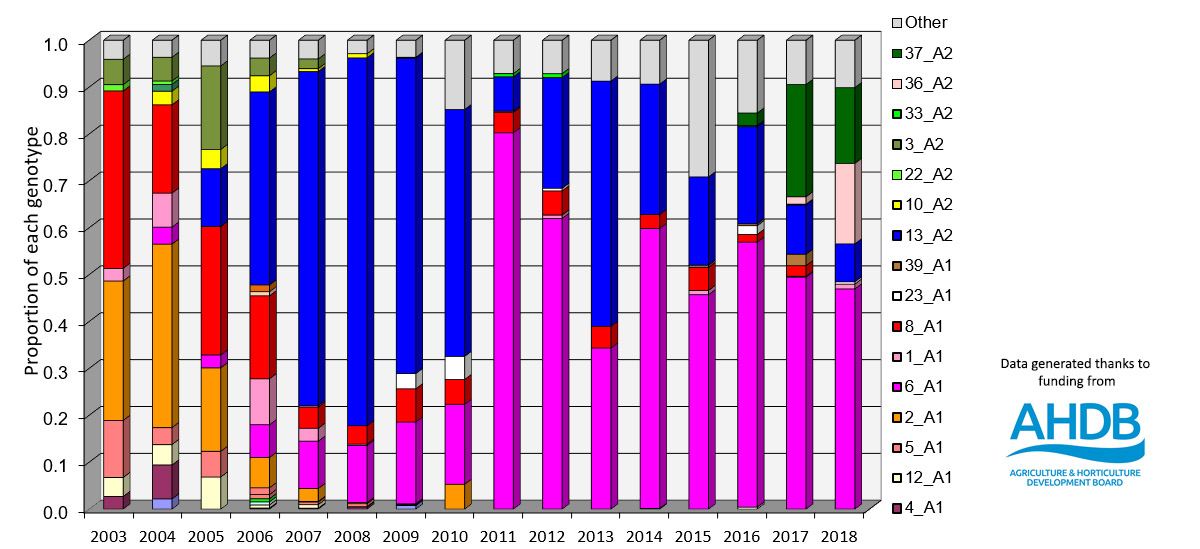
The diversity of P.infestans Genotypes in UK Fields 2003 - 2018
The Resistance
All plants possess a large family of genes that encode Resistance Proteins which help protect the plant against disease and each of these functional Resistance Proteins scans plant cells for a unique companion protein, delivered by a pathogen.
If a pathogen protein is detected by a Resistance protein, it signals for localised plant cell suicide to limit further spread of the perceived pathogen.
The resistance potential of each potato cultivar depends on the differing combination of functional resistance genes it possesses versus the varying cocktails of proteins each pathogen isolate uses to cause infection.

The challenge
The challenge for scientists and breeders is to identify and deploy the Resistance Proteins that recognise the widest range of isolates.
The potato genome has 840 million DNA base pairs but only 0.24% of these contain Resistance Proteins, equating to just 755 Resistance Genes. The function of most of these Resistance Genes is unknown.
If you picture the potato genome as the 2600km journey from Paris to Moscow, Resistance Genes only make up 6km of this journey and a single Resistance Gene is the equivalent of tiny, 8.4m, so finding them is not an easy task.
dRenSeq
dRenSeq is a revolutionary new diagnostic tool, developed by our colleagues at the James Hutton Institute that allows Resistance Gene sequences to be rapidly and confidently detected in all types of Solanum species.
Using dRenSeq, scientists and potato breeders can pinpoint varietal resistance to late blight and other major pathogens, ensuring healthier, resistant potatoes for the future.
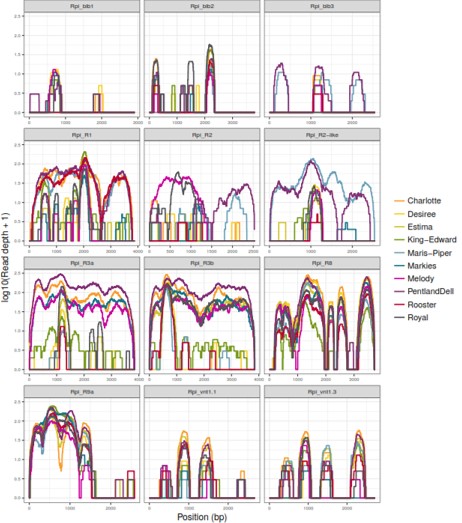
dRenSeq on the UK's 10 Most Popular Cultivars
To find out more about dRenSeq, please contact us.

